Automating RNA-Seq Downstream Analysis with DESeq2 and ClusterProfiler
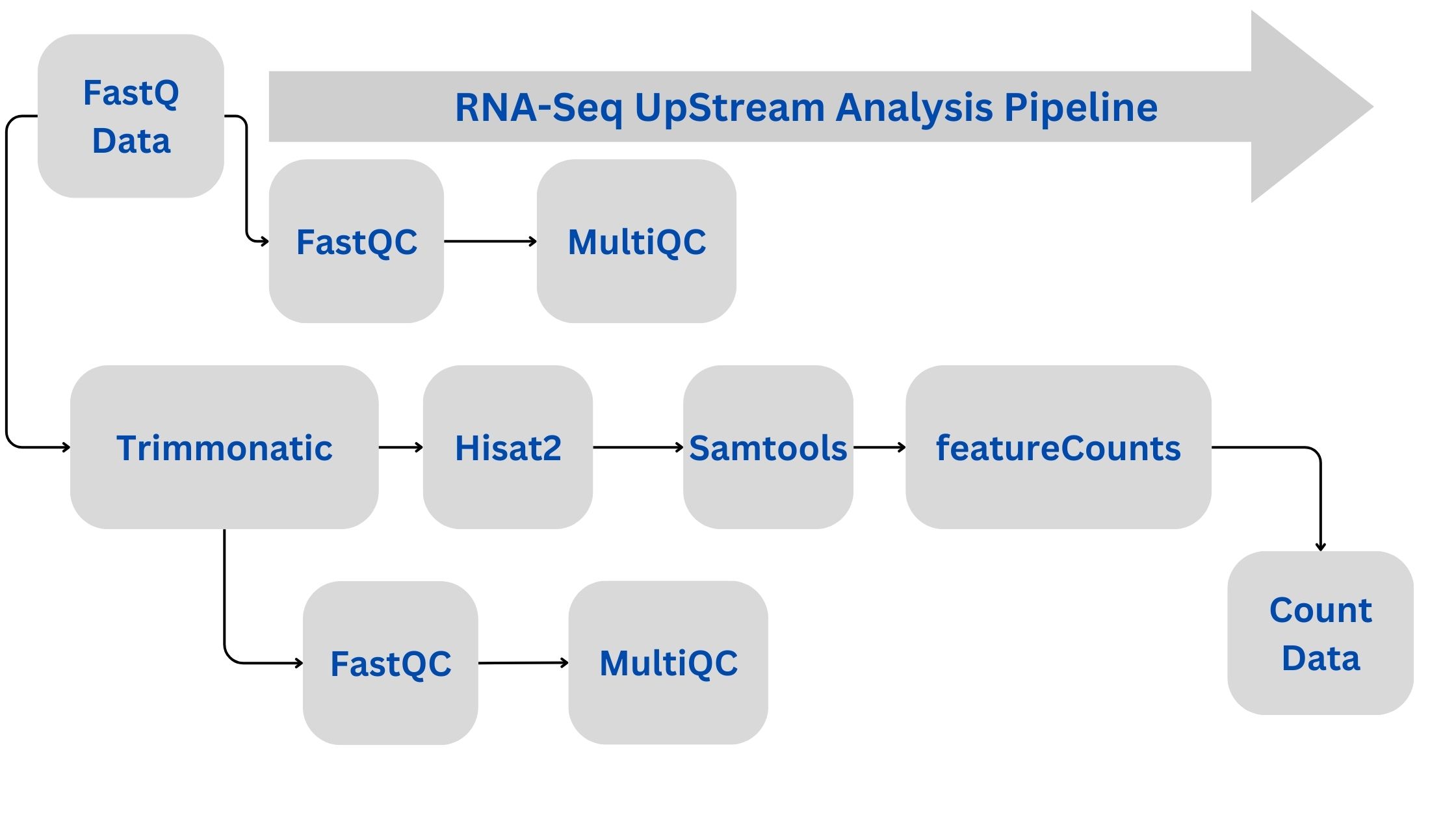
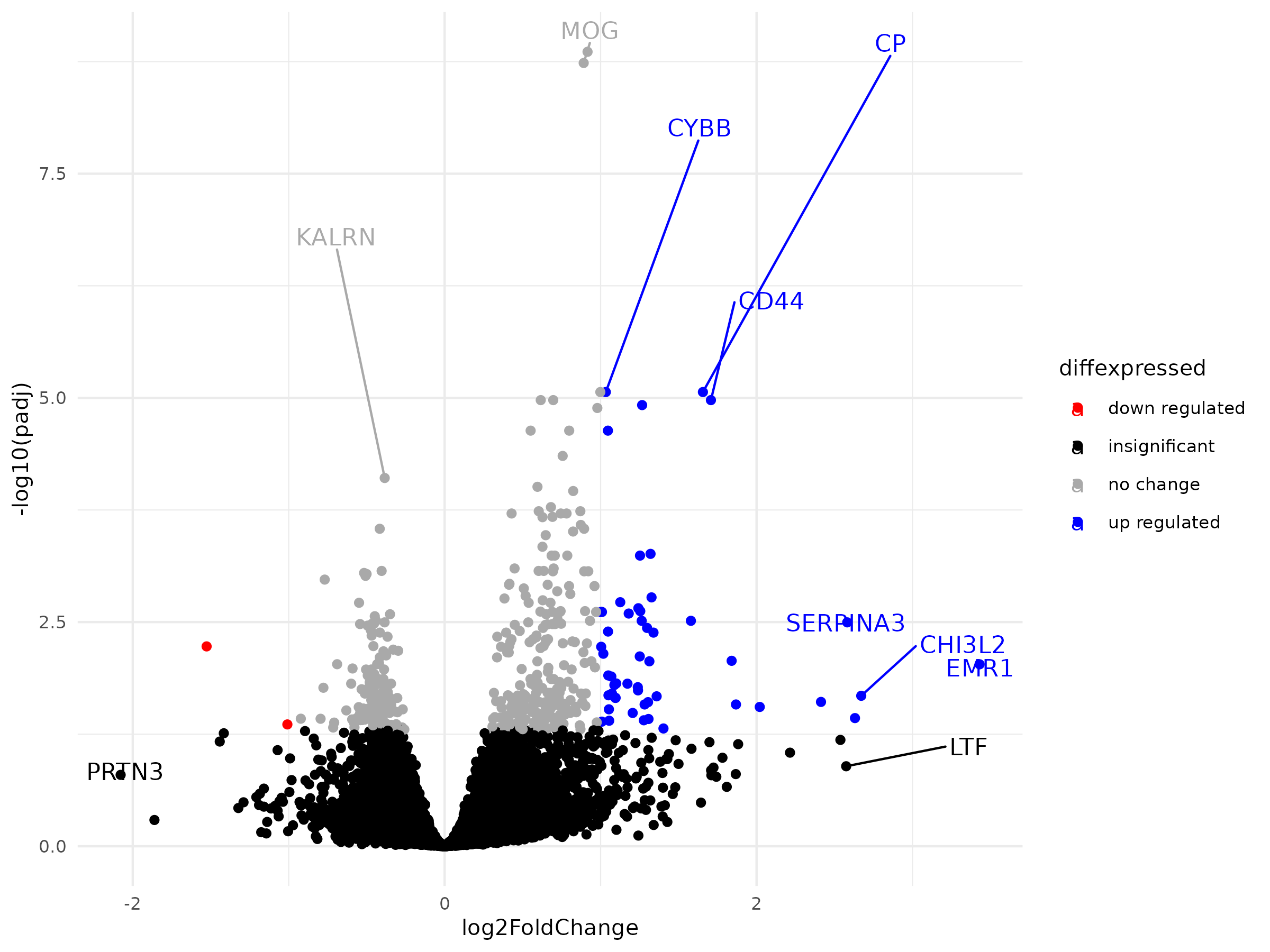
In the realm of transcriptomics, RNA-Seq has become a powerful tool for understanding gene expression patterns. However, the journey from raw data to meaningful biological insights involves several intricate steps of data processing and analysis. In this tutorial, we will walk through a comprehensive RNA-Seq downstream analysis workflow using DESeq2 and ClusterProfiler, split into distinct stages for clarity and automation.